Electrostatic Similarity¶
The electrostatic similarity method generates grid potentials for a list of PDB files and compares
all potential files in a pairwise manner. Here we will provide test cases that compares several
members of a family of plant proteins. You may download all necessary files for these examples at this link:
download. These examples are based on a more comprehensive,
published study [Chae2010].
ElecSimilarity(pdbfiles[, pdb2pqr_exe, ...]) |
Summary |
Note
The ElecSimilarity method should only be used to compare structurally and functionally similar proteins.
Additionally, all protein structure file should be superposed in a consistent grid space. While the method
implements a superpositioning algorithm from Modeller, the user should verify that the final structures are
suitably superposed. For some applications, users may aquire better results with a different superpositioning
scheme. These structure files can be found in the pdb_files or pqr_files folder within
the job directory.
Example case 1: LTP plant proteins¶
Open a new python session, import the ElecSimilarity class, and import the plotDend and plotESD functions:
from aesop import ElecSimilarity, plotDend, plotESD
Warning
If you are planning to leverage multiple CPU threads for a faster analysis, please know that extra steps may be required. Specifically, you must protect the entry point of the program according to multiprocessing documentation. You may do this by putting the following code at the beginning of your Python script:
if __name__ == '__main__':
# place remaining code here and maintain level of indentation
This precaution becomes unnecessary if you are running the analysis inside an interactive Python session. In the downloadable zip files, we have already placed this protection in the run script, so that you may run the analysis as follows in your platform’s terminal:
python run_ltp_ex1.py
Failure to protect the entry point may result in an infinite loop of process spawning.
Next, you must specify the full paths to your apbs and pdb2pqr executables, if
the paths for the directories containing the executables have not already been added to the environment.
Here is an example for a Windows system:
path_apbs = 'C:\\APBS\\apbs.exe'
path_pdb2pqr = 'C:\\PDB2PQR\\pdb2pqr.exe'
Now we will specify what PDB files the method should compare. Here we will use only 3 PDB files
(download). After downloading the PDB files, unzip them and place
them in the current working directory:
pdbfiles = ['1MZL.pdb', 'SCA1.pdb', 'SCA3.pdb']
Warning
If you are using your own PDB, make sure the PDB contains no missing heavy atoms. Consider also removing non-standard amino acids. PDBFixer is one option for cleaning PDB files in preparation for AESOP.
Note
If you only provide a single PDB file, AESOP will generate a library of mutants by side-chain truncation as in the Alascan class. You can force the ElecSimilarity class to generate mutants for all structures by specifying a list of selection strings that describe all regions of the PDB to mutate.
When the method is run, intermediate files will be generated and stored in a folder of the current working directory. The user has the option of naming this folder by specifying a job name:
jobname = 'LTP_test1'
Next, the method is initialized by:
family = ElecSimilarity(pdbfiles=pdbfiles,
pdb2pqr_exe=path_pdb2pqr,
apbs_exe=path_apbs,
jobname=jobname)
Finally, we are ready to run the analysis. To superpose structures before running, set superpose to True (please not that this superpositioning algorithm requires the Modeller library). To center structures before running, set center to True. Ideally, the end user should ensure that all PDB structures have consistent coordinates. This analysis will take several minutes, so please be patient:
family.run(superpose=True, center=False)
If you are running your analysis on a workstation and want to parallelize the calculation, then you may do so as follows:
family.run_parallel(superpose=True, center=False)
After the run is complete, AESOP will report if any Warnings or Errors were detected in APBS or PDB2PQR. The full logs are stored in the family.logs and can be viewed or written to file in the following manner:
family.viewLogs()
family.writeLogs(filename="family_logs.txt")
You can view results using built-in functions:
plotDend(family, filename='dend.png')
plotESD(family, filename='esd.png')
plotDend should produce a dendrogram similar to the following figure.
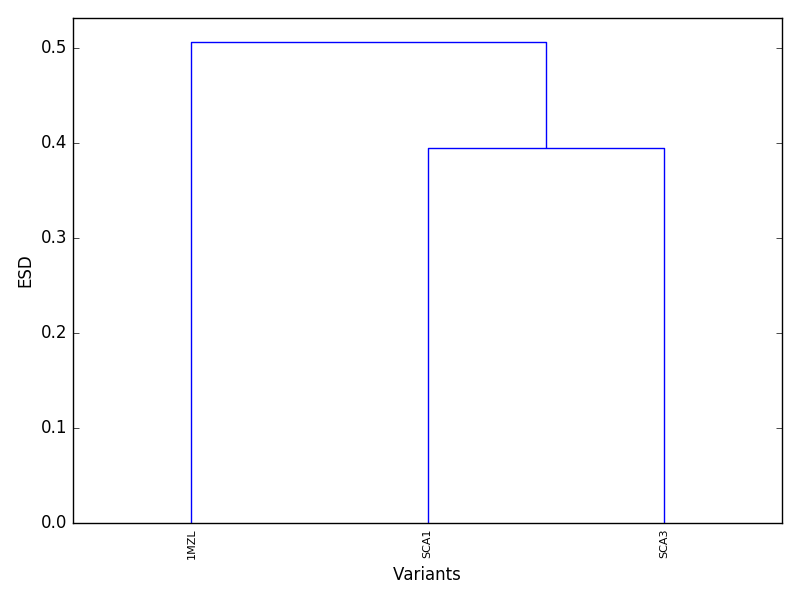
Proteins that cluster together at lower ESD in the dendrogram are expected to be electrostatically similar.
plotESD should produce a heatmap similar to the following figure.
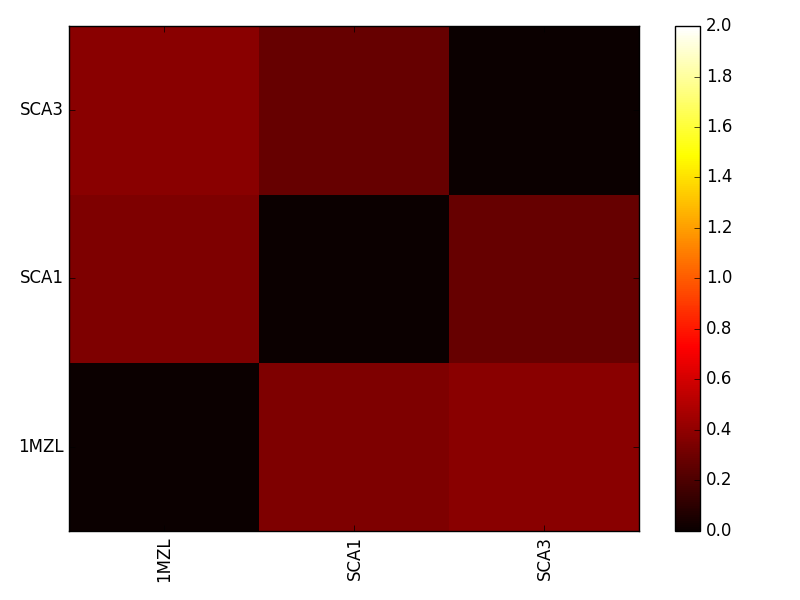
This heatmap compares all protein pairs in terms of ESD. Lower values once again indicate electrostatic similarity.
If you would like to utilize the interactive plotting function plotESD_interactive which generates a heatmap and dendrogram, refer
to this notebook demonstration.
If you prefer to export the raw data, you can access the ESD matrix:
data = family.esd
Other modules such as numpy (example below) or pandas will allow exporting of the ESD matrix to file:
import numpy as np
np.savetxt('esd_matrix.txt', data, fmt='%.4f')
The ElecSimilarity class also supports calculation of the electrostatic similarity index (ESI) by comparing potentials across all protein structures at corresponding grid points. If you previously calculated the ESD, then you need type the following to perform this calculation:
family.calcESI()
If you prefer to calculate ESI instead of ESD, you may do so at the time you run the analysis:
family.run(esi=True, esd=False, superpose=True)
family.run_parallel(esi=True, esd=False, superpose=True)
After performing the ESI calculation, you may view the ESI values by loading the DX file that is located within the “esi_files” folder of the job directory.
Example case 2: Alascan of a LTP plant protein¶
AESOP is additionally capapable of comparing electrostatic potentials of alanine mutants for a protein structure. The process follows many of the same steps from example case 1:
from aesop import ElecSimilarity, plotDend, plotESD
path_apbs = 'C:\\APBS\\apbs.exe'
path_pdb2pqr = 'C:\\PDB2PQR\\pdb2pqr.exe'
pdbfiles = ['1MZL.pdb']
jobname = 'LTP_test2'
family = ElecSimilarity(pdbfiles=pdbfiles, pdb2pqr_exe=path_pdb2pqr, apbs_exe=path_apbs, jobname=jobname)
The analysis can be run with the same run or run_parallel methods as before; however, AESOP will only know to
generate a family of alanine mutants if the length of the pdbfiles is 1 or if selstr is specified in the run statement.
selstr will tell AESOP to mutate all ionizable amino acids selected by any of the elements of the list. Only one amino
acid will be mutated at a time. The following is an example run statement that will generate mutants for all ionizable amino acids
in the 1MZL pdb file:
family.run(superpose=False, esd=True, esi=True, selstr=['protein'])